New detection technique counts COVID virus particles in saliva, breath
As health and research institutions continue to rapidly develop new methodologies for detecting SARS-CoV-2, researchers have found themselves at both forefronts of discovery and featured on the cover of the Journal of the American Chemical Society with their paper "Label-free Digital Detection of Intact Virions by Enhanced Scattering Microscopy."
Label-free detection, an approach that utilizes a biosensor and detection instrument for viral load monitoring, is a solution that can capture and digitally count intact virus particles in saliva or exhaled breath to provide lower cost and reduced time to diagnose an infection.
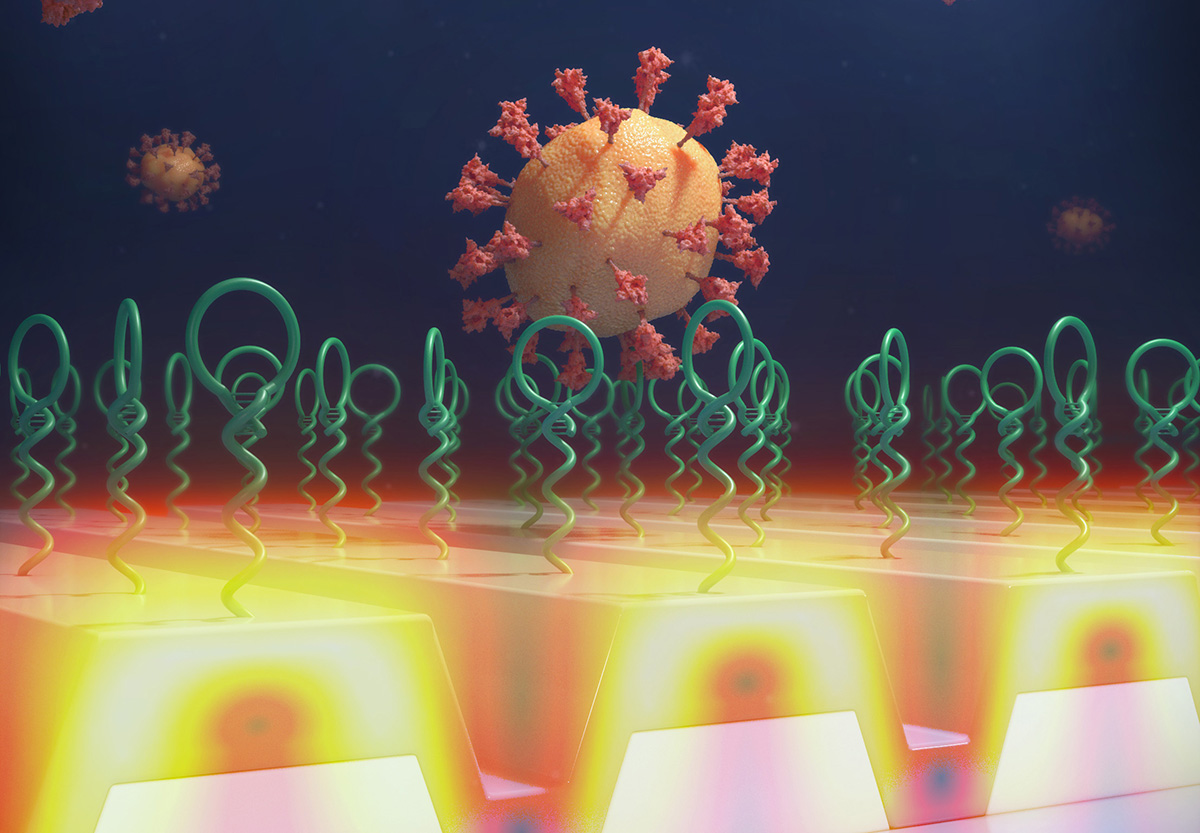
Currently, the most widely used SARS-CoV-2 PCR testing method is the PCR assay, which uses enzymatic amplification to make many copies of a specific section of the virus’s RNA, which requires extraction of the viral genome and a complex laboratory procedure. Instead, the new approach uses a specially designed nucleic acid molecule, called an “aptamer” attached to a biosensor that selectively recognizes one of the proteins on the virus outer surface, and captures it in a single step at room temperature, with no other reagents required. Once captured, the viruses are counted, using a newly invented form of microscopy, that generates images from laser light that scatters from each captured virus.
“Our technique requires only the saliva sample, and avoids the need for any additional reagents, thus we expect the cost for a test to be significantly reduced and the overall process to be greatly simplified (less labor-intensive),” said co-author Nantao Li, a graduate student in electrical and computer engineering. “In addition, the aptamers we used can selectively differentiate between active viruses from inactive ones, thus providing more robustness for diagnosis results. Conventional techniques, such as PCR, detect the viral RNA sequence which can remain in bodily fluids even after infectious viruses are no longer present.”
To detect and count the captured viruses, the team recently invented a new imaging approach called Photonic Resonator Interferometric Microscopy (PRISM). PRISM uses a photonic crystal biosensor surface to enhance light scattering from virus particles. The photonic crystal is a nanostructured surface that provides two effects. First, it enables each virus to scatter more light from an illuminating laser, which increases their signal contrast. Secondly, the photonic crystal directs the scattered light toward the microscope objective – allowing a larger fraction of the scattered light to be collected.
While label-free digital detection can be a promising alternative to traditional SARS-CoV-2 detection, principal investigator Brian Cunningham (CGD Director/MMG) believes this approach can be widely applicable to other areas.
“We are already making plans soon to perform viral load monitoring of HIV in plasma, and we are building a more portable version of the detection system that would be small and inexpensive enough to perform well in biology labs or diagnostic lab facilities," said Cunningham, Intel Alumni Endowed Chair in Electrical and Computer Engineering.
Moving forward, the research team – comprised of principal investigators Brian Cunningham, Yi Lu, Xing Wang (CGD), and co-authors Xioajing Wang, Nantao Li, and Joseph Tibbs, are designing and implementing a new type of capture molecule that will reduce the detection time to a few minutes. They envision the future capability for a person to exhale into a device, and for exhaled virus particles to be captured on the sensor.
